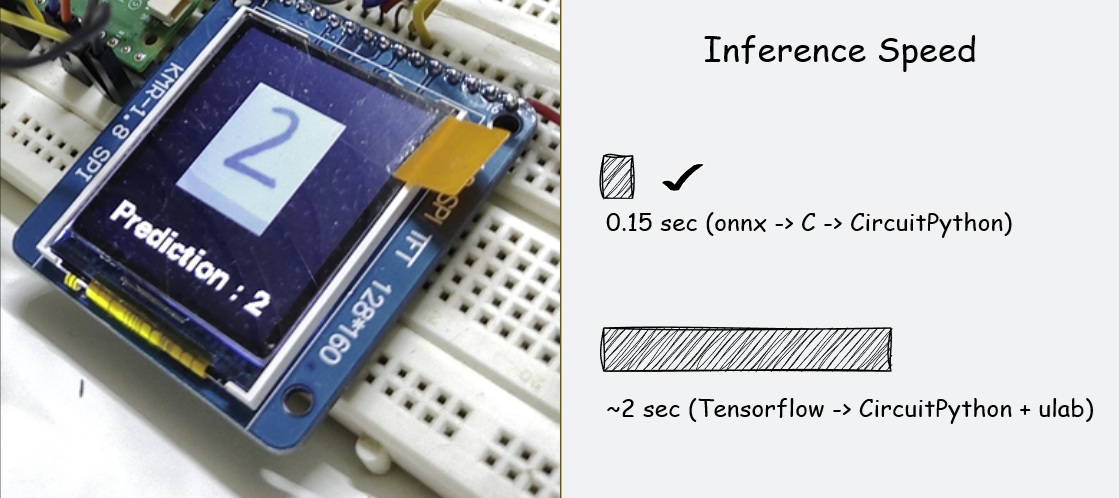
CircuitPython is loved for its simplicity, excellent documentation, and rich ecosystem of libraries. But when it comes to machine learning, especially deep learning, things quickly get complicated.
Training models? Impossible on-device.Running inference? Painfully slow if done in pure Python.
In th ...
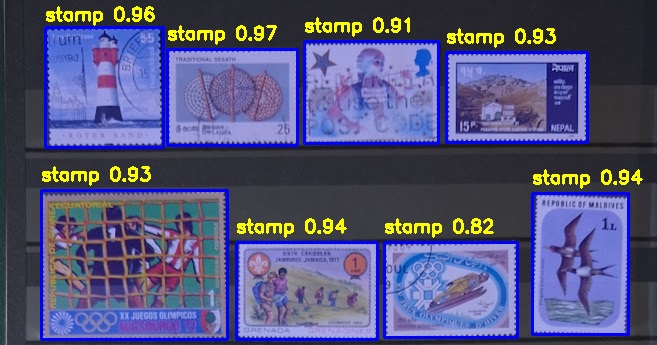
Digitizing Stamp Albums - A Lightweight, Open Source, Local AI Tool for Philatelists
Philatelists often maintain large collections of postal stamps stored across multiple albums. Over time, these collections grow into entire shelves, making it increasingly difficult to:
Keep track of what stamps are already owned
Find a specific stamp quickly
Search by visual appearance or textual ...
From Tiny Models to Private AI - Fine-Tuning SLMs and Running Them Securely in Your Web Browser
One of my long-standing interests has been making things smaller, faster, and more efficient — whether it’s embedded systems or TinyML (Tiny Machine Learning). This philosophy naturally extends to language models, where a growing movement is exploring Small Language Models (SLMs) as an efficient al ...
Domain Adaptation of Small Language Models - Fine-Tuning SmolLM-135M for High-Fidelity Paraphrase Generation
I recently embarked on an adventure to fine-tune HuggingFace’s SmolLM-135M, one of the smallest language models available. The goal was to tame this lightweight model and explore its performance on paraphrasing tasks.
Dataset ChoiceInstead of the usual Quora Question Pairs (QQP) Dataset, I chose PA ...
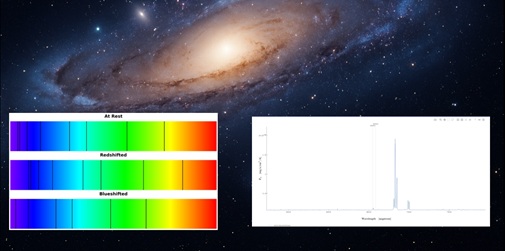
Galaxy Speed Detectives - Measure How Fast a Galaxy Moves
Have you ever looked up at the night sky and wondered: How fast are those galaxies moving? The universe is constantly expanding, and galaxies are racing through space at unimaginable speeds. Astronomers use a technique called redshift and blueshift to measure whether galaxies are moving toward ...
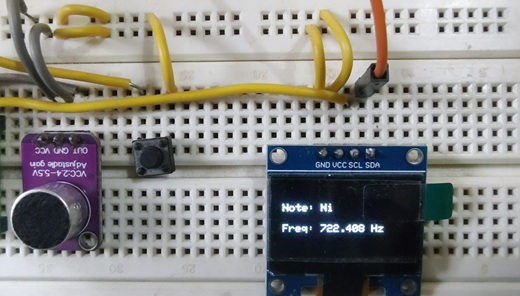
Identifying Flute Notes Using CircuitPython, FFT, and a Budget Microphone
The Raspberry Pi Pico never ceases to amaze! I love pushing CircuitPython to its limits. In this article, we dive into using CircuitPython, Fast Fourier Transform (FFT), and a budget-friendly microphone breakout board to recognize flute notes and write them on an OLED screen.
Video of the projec ...
Creating a CNN to Classify Cats and Dogs for Kendryte K210 Boards
When working with Kendryte K210 boards, a cost-effective option for edge AI projects, you’ll quickly notice that the documentation is often limited. After spending significant time researching and experimenting, I decided to document my process to help others save time when building a Convolutional ...
Training a Custom Model for a Kendryte K210 Board - Key Insights
For some time, I’ve been deeply engrossed in the challenge of training a custom model for my Maix Dock M1, a K210 board. This journey hasn’t been without its frustrations, especially due to the lack of comprehensive documentation, which turned out to be a significant hurdle. However, after a lo ...
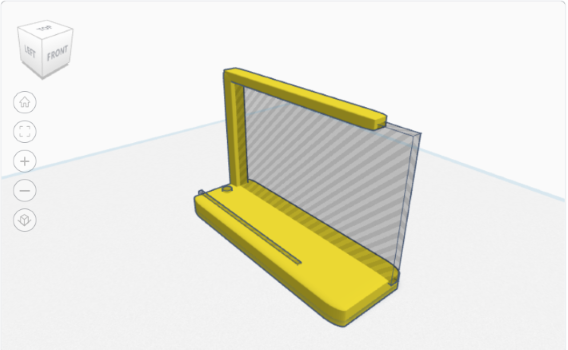
A Custom 3D-Printed Stand for Maix Dock M1
I recently bought a Maix Dock M1, which is a neat little device with a K210 neural network accelerator. The kit also included a small LCD display, which I was excited to try out. However, there was one problem—there was no case for the mainboard and the LCD. Handling the camera and the LCD togethe ...
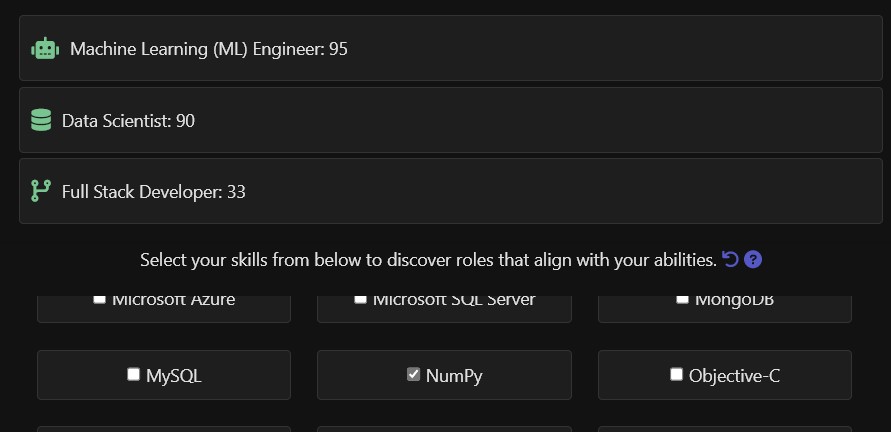
A tool to discover software career paths based on skills
When I engage with students and aspiring job seekers, a common question arises: “What roles can I pursue in the software industry?” or “What skills are needed to become ‘X’ in a software company?”
To address these queries, I have developed a user-friendly application. By visiting this url, you can ...